In Salesforce Einstein, we use Apache Spark to perform parallel computations on large sets of data, in a distributed manner. In this article, we will take a deep dive into how you can optimize your Spark application with partitions.
Introduction
Today, we often need to process terabytes of data per day to reach conclusions. To do this in an acceptable timeframe, we need to perform certain computations on that data in parallel.
Parallelizing on one machine will always have limits, no matter how big the machine is. Our true factor of parallelism is determined by the number of cores on our machine (128 is the maximum today on a single machine). So instead, we want to be able to distribute the load to multiple machines. That way, we can use a fleet of commodity hardware and reach an “infinite” parallelism factor (we can always add new machines).
Spark is a distributed processing system that helps us distribute the load on multiple machines, without the overhead of syncing them and managing errors for each. But the thing is, it’s not Spark’s job to decide how best to distribute the load. It keeps default configuration to help us get started, but those are not enough — relying on the defaults can lead to a 70% performance gap when not tuning our application, as we will see in our example later on. Our job is to tell Spark exactly how we want to distribute the load on our dataset. To do so, we must learn and understand the concept of partitions.
What is a partition?
Let’s start off with an example. Say we have a table with three columns: class ID, student ID and grade. We want to calculate the average grade for each class ID. While calculating each class average grade is independent from other classes, grades within each class are dependent on each other to reach the average grade of that class. Thus, here we can parallelize our program to the maximum number of Class IDs, but no more than that. In Spark, this is called a “partition,” and in our example we have partitioned our data by a certain column — Class ID. Computations on datasets in Spark are translated into tasks where each task runs on exactly one core. Each task (you guessed it) maps to a single partition, so the number of tasks equals the number of partitions.

Not all computations are born equal
Spark works in a staged computation; it bundles certain computations into the same stage and executes them with the same level of parallelism.
To understand how Spark decides what to bundle into a stage, we need to understand the two kinds of transformations that we can perform on our data:
- Narrow transformations: These are transformations in which data in each partition does not require access to data in other partitions in order to be fully executed. For example, functions like map, filter, and union are narrow transformations.
- Wide transformations: These are transformations in which data in each partition is not independent, requiring data from other partitions in order to be fully executed. For example, functions like reduceByKey, groupByKey, and join are wide transformations.
Wide transformations require an operation called “shuffle,” which is basically transferring data between the different partitions. Shuffle is considered to be a rather expensive operation, and we should avoid it if we can. Shuffle will result in different partitions.
Notice that often when we have x amount of partitions and we are doing a wide transformation (i.e. groupBy), Spark will first groupBy in each initial partition and only then shuffle the data, partition it by key, and groupBy again in each shuffled partition, to increase efficiency and reduce the amount of rows while shuffling.
In Spark, each stage is built from transformations that can be done in a row, without the need for shuffle (narrow transformations). To recap, stages are created based on chunks of processing that can be done in a parallel manner, without shuffling things around again.

Controlling the number of partitions in each stage
As mentioned before, Spark can be rather naive when it comes to partitioning our data correctly. That’s because it’s not really Spark’s job. We should make sure our data is well-partitioned in each executing stage.
The adaptive query execution (introduced in Spark 3) is now enabled by default in the latest Spark version (3.2.1), and you should keep it that way. Among other things (including determining join strategies in real-time and optimizing skewed joins), it reduces the number of partitions used in shuffle operations in run-time, as a dependent of dataset size (hence, adaptive). But, those are still pre-run static config values that we have to set appropriately.
Let’s have a look at some useful configuration values regarding Spark partitions:
spark.sql.files.maxPartitionBytes: The maximum number of bytes to pack into a single partition when reading files. Default is 128 MB.spark.sql.files.minPartitionNum: The suggested (not guaranteed) minimum number of partitions when reading files. Default isspark.default.parallelismwhich equals to two or the total number of cores in our cluster, whichever is bigger.
spark.sql.adapative.enabled and spark.sql.adaptive.coalescePartitions.enabled must be true (which is the default case) for the next config values to apply:
spark.sql.adaptive.advisoryPartitionSizeInBytes: Target size of shuffle partitions during adaptive optimization. Default is 64 MB.spark.sql.adaptive.coalescePartitions.initialPartitionNum:As stated above, the adaptive query execution optimizes while reducing (or in Spark terms – coalescing) the number of partitions used in shuffle operations. This means that the initial number must be set high enough. This value is the initial number of partitions to use when shuffling, before starting to reduce it. The default isspark.sql.shuffle.partitions, which equals to 200.spark.sql.adaptive.coalescePartitions.parallelismFirst:When this value is set to true (the default), Spark ignoresspark.sql.adaptive.advisoryPartitionSizeInBytesand only respectsspark.sql.adaptive.coalescePartitions.minPartitionSizewhich defaults to 1M. This is meant to increase parallelism.
If we set spark.sql.adapative.enabled to false, the target number of partitions while shuffling will simply be equal to spark.sql.shuffle.partitions.
In addition to to these static configuration values, we often need to dynamically repartition our dataset. One example is when we filter our dataset. We might end up with uneven partitions, which will cause skewed data and un-balanced processing. Another example could be when we want our data to be written in a partitioned way to different folders by certain key. We might want our dataset partitioned by that key in memory beforehand, to avoid searching in multiple partitions while writing.
Having said that, let’s see how we can dynamically repartition our dataset using Spark’s different partition strategies:
- Round Robin Partitioning: Distributing the data from the source number of partitions to the target number of partitions in a round robin way, to keep equal distribution between the resulted partitions. Since repartitioning is a shuffle operation, if we don’t pass any value, it will use the configuration values mentioned above to set the final number of partitions. Example of use:
df.repartition(10).

- Hash Partitioning: Splits our data in such way that elements with the same hash (can be key, keys, or a function) will be in the same partition. We can also pass wanted number of partitions, so that the final determined partition will be hash % numPartitions. Notice that if numPartitions is bigger than the number of groups with the same hash, there would be empty partitions.
Example of use:df.repartiton(10, 'class_id')

- Range Partitioning: Very similar to hash partitioning, only it’s based on a range of values. Due to performance reasons, this method uses a sampling to estimate the ranges. Hence, the output may be inconsistent, since the sampling can return different values. The sample size can be controlled by the config value
spark.sql.execution.rangeExchange.sampleSizePerPartition.
Example of use:df.repartitionByRange(10, 'grade')

When decreasing our partitions, we can also use the coalesce method, which does not perform data shuffling. It simply merges existing partitions (as opposed to repartition, which shuffles the data and creates new partitions). Hence, it is more performant than repartition.
But, it might split our data unevenly between the different partitions since it doesn’t uses shuffle. In general, we should use coalesce when our parent partitions are already evenly distributed, or if our target number of partitions is marginally smaller than the source number of partitions. In other cases, we would probably want to use repartition, to make sure our data is distributed evenly.
What is the optimal number of partitions?
Of course, there is no one answer to this question. How you should partition your data depends on:
- Available resources in your cluster
Spark’s official recommendation is that you have ~3x the number of partitions than available cores in cluster, to maximize parallelism along side with the overhead of spinning more executors. But this is not quite so straight forward. If our tasks are rather simple (taking less than a second), we might want to consider decreasing our partitions (to avoid the overhead), even if it means less than 3x. We should also take under consideration the memory of each executor, and make sure we are not exceeding it. If we do, we might want to create more partitions, even if it’s more than 3x our available cores, so that each will be smaller, or increase the memory of our executors. - How your data looks (sizing, cardinality) and computations being made
You should make sure that your data is distributed as evenly as possible across your different partitions. Sometimes it can be easily done (using round robin partitioning), but other times it can be more complicated. Group by, windowing, and writing our data in a way that it will be easily queried later often require partition by key. When doing this, we need to make sure that one group’s values are not marginally larger than the others. If that’s the case, we need to see if we can achieve the same result with different measures by normalizing our data or changing our computations (i.e. split this specific key into two, apply certain computations, and then merge between them).
Spark Partitions in Action
Let’s put some of what we’ve learned about Spark partitions into an example.
Say we have a dataset weighing ~24 GB that contains fraud deals that occurred in 2021, in certain businesses.
It looks like this:

For each business, we want to calculate the total amount of fraud deals made in that business and how this total amount compares to the average total amount.
In the end, we want to write the result as csv — partitioned to folders by total business fraud amount/average fraud amount (accurate to the third decimal point).
Let’s see a naive example of how to do it using pyspark and Spark’s DataFrame API:
spark = SparkSession.builder.getOrCreate()
df = spark.read.option('header', 'true').csv('./example_data/dataset_1.csv')df = df.withColumn('amount', F.col('amount').cast('int'))
df = df.groupBy('business').agg(F.sum('amount').alias('total_amount'))df_avg = df.select(F.avg('total_amount').alias('avg_amount'))
df = df.crossJoin(df_avg)
df = df.withColumn('compared_to_avg', \
F.round(F.col('total_amount') / F.col('avg_amount'), 3))
df.write.mode('overwrite').partitionBy('compared_to_avg').csv('./output_data/')Great! We’ve done it. The main stages in our application are:
- Cast amount to int and group by business in each partition to partially sum the total amount of each business (partially because this business can also appear in other partitions)
- Shuffle the data so that it is partitioned by business, summing the total amount for each business and calculating the average amount for each business
- Create another DataFrame that contains the average total amount (of all business) and join both data frames to add the average amount for each row
- Calculate compared_to_avg as the total business fraud amount / average fraud amount and write again as csv, partitioned by compared_to_avg
Now let’s look at some statistics in Spark UI.
Stage #1:

Spark used 192 partitions, each containing ~128 MB of data (which is the default of spark.sql.files.maxPartitionBytes).
The entire stage took 32s.
Stage #2:

We can see that the groupBy shuffle resulted in 11 partitions, each containing ~1 MB of data (which is the default of spark.sql.adaptive.coalescePartitions.minPartitionSize).
The entire stage took 0.4s.
Stage #3:

Spark used 1 partition containing 708 B to fully calculate the average total amount and join this data with each row.
The entire stage took 4ms.
Stage #4:
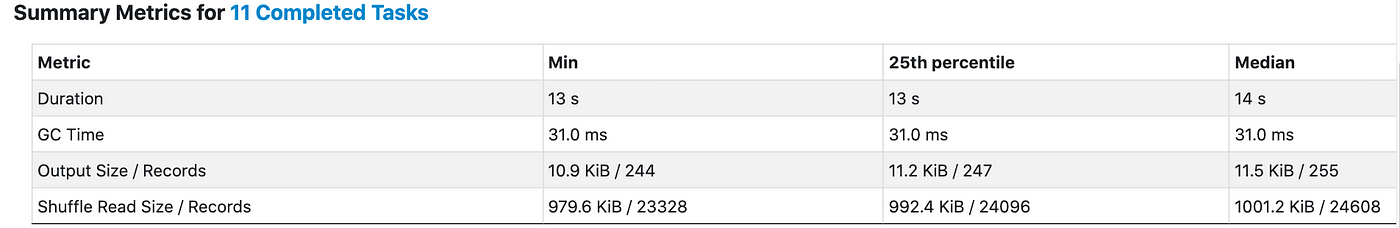
We can see that join shuffle resulted back in 11 partitions, each containing ~1 MB of data (which is the default of spark.sql.adaptive.coalescePartitions.minPartitionSize).
The entire stage took 15s.
So… how can we optimize it? First, we need to have knowledge of our data size, it’s cardinality, and our resources. We already know that our dataset weighs ~24 GB. Regarding its cardinality, we can see that after we grouped our dataset by business, its size was down by a lot (to around 11 MB), meaning that we have a low cardinality when looking at the different businesses. Regarding our resources— I’m running Spark in local mode, and my local machine has 16 cores and 32 GB of RAM.
As mentioned in the previous section, to maximize parallelism, we want 3x partitions. So, for stage #1, the optimal number of partitions will be ~48 (16 x 3), which means ~500 MB per partition (our total RAM can handle 16 executors each processing 500 MB). To decrease the number of partitions resulting from shuffle operations, we can use the default advisory partition shuffle size, and set parallelism first to false. (Spark documentation also recommends you set this value to false when you know what you’re doing.) One last thing— before writing, our dataset is partitioned by business (as a result of our groupBy), but we write our data partitioned by compared_to_avg column. It might be a good idea to partition it on compared_to_avg before writing.
Let’s see how the optimized version looks:
spark_conf = SparkConf()
spark_conf.set('spark.sql.adaptive.coalescePartitions.initialPartitionNum', 24) spark_conf.set('spark.sql.adaptive.coalescePartitions.parallelismFirst', 'false')
spark_conf.set('spark.sql.files.minPartitionNum', 1)
spark_conf.set('spark.sql.files.maxPartitionBytes', '500mb')spark = SparkSession.builder.config(conf=spark_conf).getOrCreate()df = spark.read.option('header', 'true').csv('./example_data/dataset_1.csv')
df = df.withColumn('amount', F.col('amount').cast('int'))
df = df.groupBy('business').agg(F.sum('amount').alias('total_amount'))
df_avg = df.select(F.avg('total_amount').alias('avg_amount'))
df = df.crossJoin(df_avg)
df = df = df.withColumn('compared_to_avg', \
F.round(F.col('total_amount') / F.col('avg_amount'), 3))
df = df.repartition(24, 'compared_to_avg')
df.write.mode('overwrite').partitionBy('compared_to_avg').csv('./output_data/')Stage #1:

Like we told it to using the spark.sql.files.maxPartitionBytes config value, Spark used 54 partitions, each containing ~ 500 MB of data (it’s not exactly 48 partitions because as the name suggests – max partition bytes only guarantees the maximum bytes in each partition).
The entire stage took 24s.
Stage #2:

We have set spark.sql.adaptive.coalescePartitions.parallelismFirst to false, and now Spark AQE uses the default value for shuffle coalescing advisory partitions size (128 MB), resulting in 1 task containing 2 MB. Notice that data size in this stage is much smaller than the previous execution, because we had fewer partitions in stage #1, and the partial groupBy in each partition resulted in much less data to shuffle and fully groupBy.
The entire stage took 0.2s.
Stage #3:

The previous stage already calculated the entire average amount for all business, because they all were in the same partition, so all this stage had left to do was simply join the output.
The entire stage took 2ms.
Stage #4:

We have another stage added — partitioning the data using the hash partitioner. Spark used 1 partition containing 2.6 MB of data.
The entire stage took 0.1s.
Stage #5:

We wanted our data partitioned into 24 partitions by compared_to_avg column, and that’s exactly what we’ve got. I chose 24 after trial and error, trying to balance between increasing the parallelism factor and the parallelism overhead (resulting in each task taking ~2s). Each of the 24 partitions held ~5.1 KB of data. The entire stage took 4s.
- Application without partition tuning: 47.4s.
- Application with partition tuning: 28.1s.
Notice the difference! We’ve managed to achieve the same goal, but much faster. By understanding the concept of partitions, how Spark manages them, and by considering the different factors, we improved our performance by 40%!
*Dataset was taken from kaggle.com.
Final note
We have learned how partitions allow us to parallelize computations on our dataset and how we can control the way Spark partitions our data, taking into consideration our dataset, computations being made, and cluster size. We’ve also learned how we can leverage all of that to make our application more performant.
While we demonstrated optimization using correct partitioning, there are other factors for tuning and optimizing our Spark application. SQL hints (i.e. for specific join strategies), dynamic repartition to avoid skewed joins, Caching, and Dynamic Resource Allocation, can all be used to optimize our application, and I encourage you to dive deeper. There’s no better place to start than the official documentation: https://spark.apache.org/docs/latest/sql-performance-tuning.html.